class: center, middle, inverse, title-slide # Combining genetic data and DNA methylation in START project ### Julia Romanowska ### 16-10-2020 --- class: left, inverse, bottom # OVERVIEW *** ## Ideas ## Examples ## Discussion ??? As you probably know, our group here loves to make things more complicated than all the others, so what we wanted to focus on is not only methylation data, but the integration of *genetics* and *epigenetics*. I will present some of our ideas and show a few examples of how the results might look like, but I'd like to hear your thoughts about these planned analyses because there are lots of discussion points and questions, and not so many answers yet. --- class: left, inverse, bottom # IDEAS *** ## meQTLs & PoO-effect ## interaction effect: PoOxMe ## X-chromosome analysis ??? These are three areas we want to focus on: - methylation quantitative trait loci, so are there any associations between methylation levels of certain CpGs and the alleles on a SNP; and are these parent-of-origin specific? - does methylation level interact with parent-of-origin genotype effect? - and finally, since all the other analyses are focused on autosomal chromosomes, we want to look at the X and Y chromosomes --- class: left, inverse, bottom ## meQTLs *involved: Julia, Miriam, Håkon, Anil, Rolv Terje* --- ## meQTLs - idea * calculate meQTLs in parents only (cases and controls separately) * is there a difference between cases and controls? <br> * calculate the same in children and check significance * is there difference between case-families and control-families? ??? We will focus first on checking SNP-CpG pairs in parents, to check simply whether there is a difference in patterns in ART-parents and non-ART-parents. This could point to specific epigenetic mechanisms that might be causal for fertility problems. Next, we can calculate meQTLs also in children and with that the number of comparisons we can make grows significantly. Each of the comparisons also gives specific interpretation. Håkon has made a very nice illustration of some of the possibilities. --- **meQTL:** No **ART-effect:** No 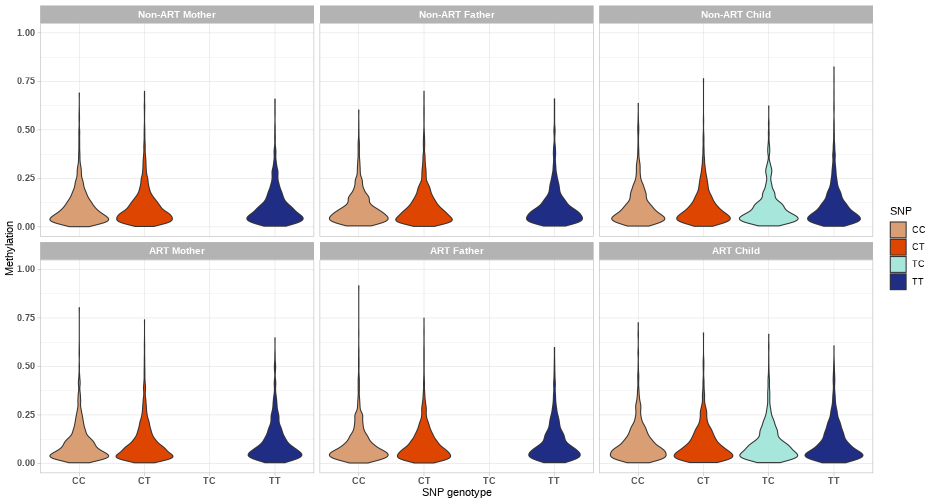<!-- --> ??? Here, basically, nothing happens. --- **meQTL:** No **ART-effect:** No 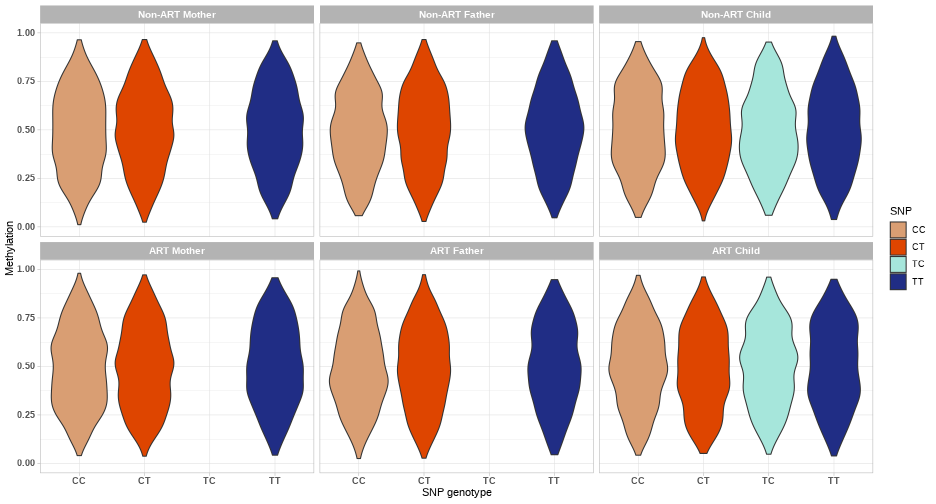<!-- --> ??? Similarly here. --- **meQTL:** No **ART-effect:** Yes 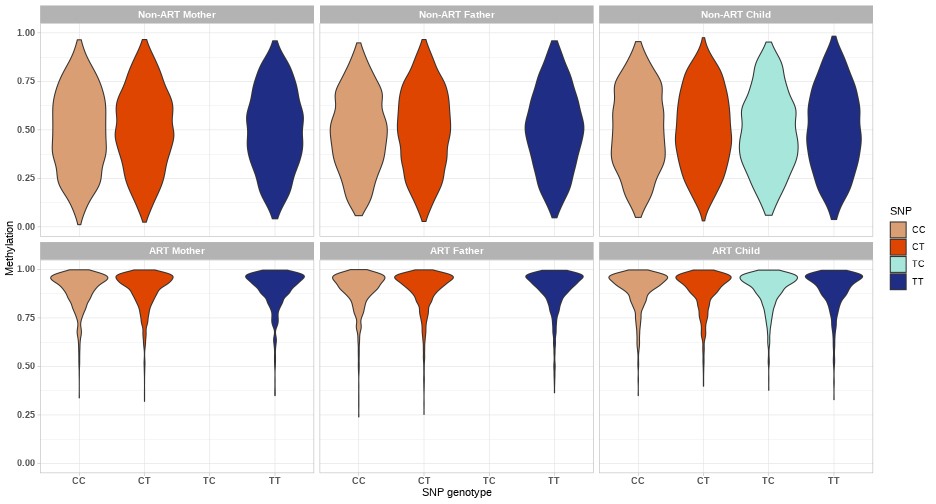<!-- --> ??? And suddently - wow, there is an effect when we compare ART-families with non-ART-families! But still no meQTL. --- **meQTL:** Yes **ART-effect:** No 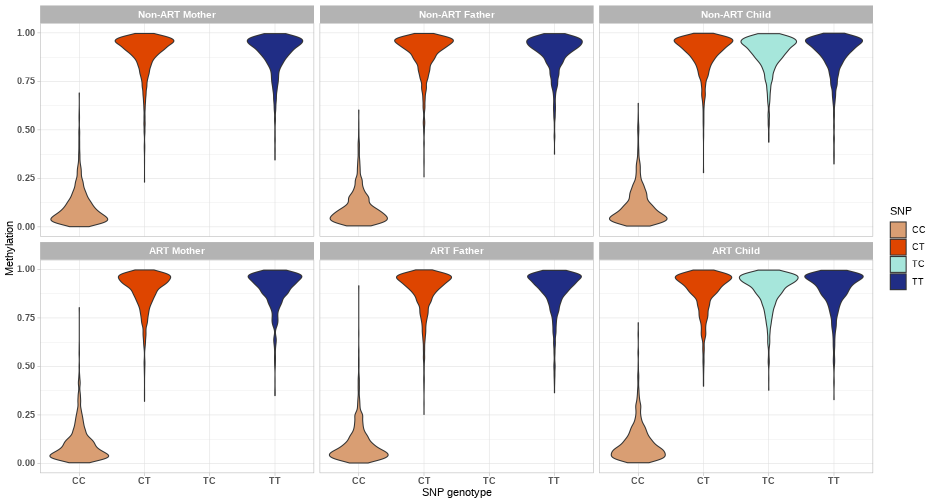<!-- --> ??? And vice-versa: the methylation now depends on genotype, but the effect is the same in ART vs. non-ART families. --- **meQTL:** Yes **ART-effect:** Yes (after fertilization?) <!-- --> ??? And suddenly, bang! Methylation is dependent on the genotype **and** the effect differs when the child is in the ART-family. But that's not all - as I've mentioned, we like comlicated things... --- **meQTL:** Yes **ART-effect:** Yes **POO ART-effect:** Yes (maternal line?) <!-- --> ??? Bam - now, there is meQTL dependent on ART-status **and** parent-of-origin specific! --- **meQTL:** Yes **ART-effect:** Yes **POO ART-effect:** Yes (paternal line?) <!-- --> --- ## meQTLs - Discussion * are meQTLs PoO-specific? *(if the ART procedures affect the methylation before fertilisation, then we should see the PoO-specific meQTLs)* * what does it say that there is (no) difference between cases and controls? * how likely it is that meQTLs are shared between child and parent? *(how do they work during the fertilisation/embryogenesis?)* * are meQTLs enriched in CpGs associated with ART? ??? With that in mind, there are tons of questions to ask! --- class: left, inverse, bottom ## interaction effect: PoOxMe *involved: Julia, Ellisif?, Håkon, Anil, Rolv Terje* ??? Anybody else? --- ## PoOxMe * use our new methodology! * Haplin & HaplinMethyl * how to treat the meQTLs found before? --- class: left, inverse, bottom ## X-chromosome analysis *involved: Julia, Haakon, Håkon, Anil, Rolv Terje* ??? Anybody else? --- ## X-chrom - idea * X-EWAS? *we need to find the right method* * what has been already done? ??? Need to check what type of information is out there: - any X-chrom EWAS? - there are also some regions of X-chrom that are not gender-dependent... - how to interpret this?